Name
Supervisor de Marie Curie: “MagMA” Applying Metabolomics to Unveil follow-up treatment biomarkers and Identify Novel Therapeutic Targets in Glioblastoma
Principal Investigator
Coral Barbas
Financial entity
MSCA-IF-2017 Project, Grant Agreement Number 799378 
Participants
1
Duration
2018-2020
Summary
Glioblastoma multiforme (GBM) is a malignant tumour originating from glial cells. It is the most common and devastating form of malignant brain tumour, containing self-renewing, tumorigenic cancer stem cells (CSCs) that contribute to tumour initiation and therapeutic resistance. It leads to 225,000 deaths per year in the entire world. Standard treatment consists of maximal surgical resection, followed by radiotherapy with or without concomitant and adjuvant Temozolomide. Such treatment hardly increases patient survival and leads to a median overall survival of only 12–18 months.
By contrast, to other types of cancers, it appears uncertain that GBM incidence can be decreased by changing certain environmental factors, or anticipated from the presence of another disease or condition. Until date, all efforts that have been done to improve patient’s life have shown low efficacy and the survival of such patients has not improved much in the last 60 years. Therefore, it seems clear that new methods for diagnosis, prognosis and treatment are needed.
To better understand GBM tumour biology, several groups worldwide have turned to high dimensional profiling studies. On 2018 Verhaak and collaborators described several GBM phenotypes; Proneural, Neural, Classical and Mesenchymal, each with distinguishing hallmark mutations, copy number alterations, epigenetic alterations, and clinical features. Additionally, treatment efficacy differs per subtype. Nevertheless, a full metabolomic profile had not been performed until date.
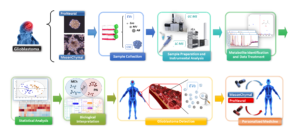
The first objective of our study was to investigate the underlying metabolic differences between the most extreme phenotypes (Proneural and Mesenchymal). These generate a characteristic fingerprint, not yet reported until now, thereby providing a better understanding of glioma biology. This finding could help in the development of new strategies to fight the tumour and lead to more personalized therapy.
Another difficulty of GBM is that both, confirmation and follow up of the tumour process are restricted by anatomical location. Nevertheless, neural cells are able to release extracellular vesicles (EVs), which cross the blood-brain barrier and could be detected within the blood, offering a potential new way for detection and treatment monitoring. The second objective of the project was to study the metabolic profile of the EVs released by those subtypes. This research effort has helped to draw the EVs metabolome and to elucidate whether or not the metabolites are directly packaged into specific EVs and their possible function in the surrounding cells. Moreover, it gives the opportunity of finding a metabolite profile characteristic of tumour subtype that could be used as a biomarker.
Dissemniation activities
Conference talks
- Application of metabolomics in the identification of new therapeutic targets in glioblastoma, as biomarkers for its treatment and evolution. Oral communication. CEU San Pablo Interuniversity Researchers Conference 31st January 2020 (D 6.2)
- Effects of mutations in the post-translational modification sites on the trafficking of hyaluronan synthase 2. Poster presentation. International Conference of Extracellular Vesicles 2019 at Kyoto, Japan on 24th-28th April. https://www.isev.org/page/ISEV2019
Public engagement activities
Outreach activities
- What it means to be a woman researcher. February 11, 2020 Alicante, Spain. The researcher has participated giving a talk in the activities organized by the Secondary School “IES la Melva” during the International day of Women and Girls in Science: https://www.instagram.com/p/B8b0B95ipmm?igshid=o1y4ltm4irpl
- A day in a laboratory of Cellular and Molecular Biology. Workshop. 19th Week of Science at the Universidad San Pablo-CEU (USP): https://www.uspceu.com/investigacion/cultura-cientifica/semanaciencia and https://www.madrimasd.org/semanacienciaeinnovacion/ (general public and School Children’s). November 4-7, 2019, Madrid, Spain.
- Marie Skłodowska-Curie Actions. European Researcher’s Night. Organized by Fundación Para El Conocimiento Madri+d. September 27, 2019. Madrid, Spain.
- Microorganisms that make you love them and microorganisms that make you fear them. Workshop. 18th Week of the Science at USP: https://www.ceuuniversities.com/en/ universidad-ceu-san-pablo-takes-part-in-the-18th-week-of-science-and-innovation/. November 5-18, 2018, Madrid, Spain.
- “Marie Skłodowska-Curie Actions”. European Researcher’s Night. Organized by Fundación Para El Conocimiento Madri+d: https://www.madrimasd.org/lanochedelosinvestigadores. September 28, 2018 Madrid, Spain.
- Participation at the event Europe’s Beating Cancer Pan in Brussels on 4th February 2020. https://ec.europa.eu/health/non_communicable _diseases/events/ev_20200204_en


Newspaper Interviews
University website News

Other activities
Teaching activities
- Dr Melero has contributed to teaching activities participating at Genetics grade in 2018-2019 and 2019-2020 academic years teaching the subject “Instrumental techniques”, and “Instrumental and Bioanalytical techniques” at bilingual grade of Biotechnology at 2018-2019 academic year, with a total of 60h.
Supervision of students
- The researcher has supervised Bethany Pembridge, a one year Erasmus student to conduct her own research project at “Universidad CEU-San Pablo”.